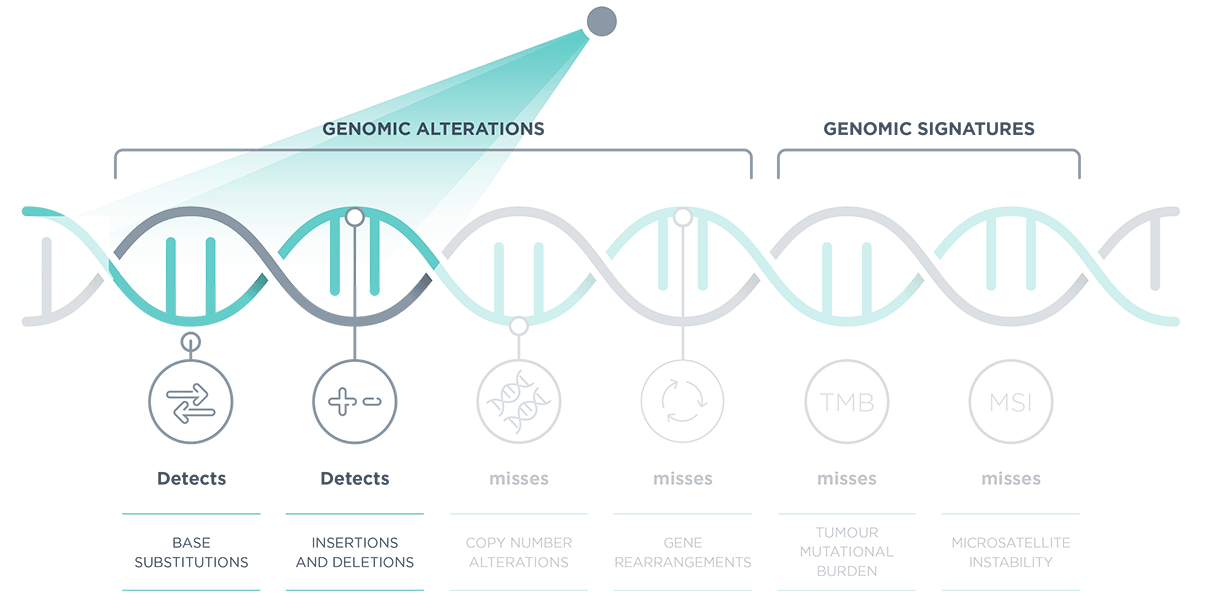
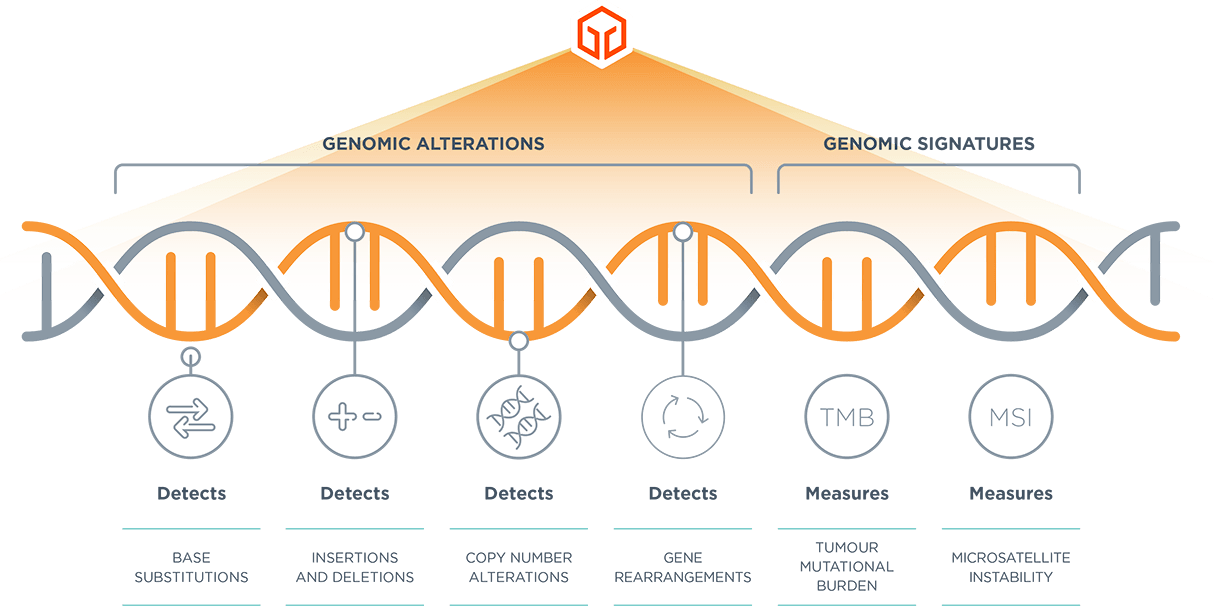
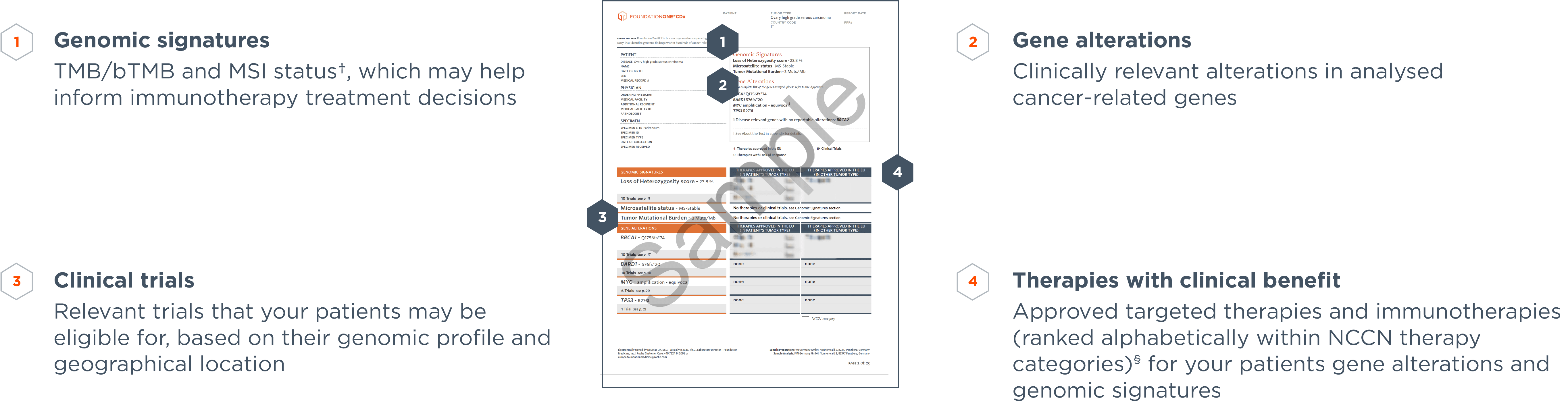
*Comprehensive genomic profiling provides prognostic, diagnostic and predictive insights that inform treatment decisions for individual patients across all cancer types.
†TMB reported by FoundationOne CDx and FoundationOne Heme. bTMB reported by FoundationOne Liquid CDx. MSI reported by FoundationOne CDx and FoundationOne Heme, MSI-H reported by FoundationOne Liquid CDx.
‡Therapies contained in the EU version of the report may have been approved through a centralised EU procedure or a national procedure in an EU Member State.
§For additional information on the NCCN categories please refer to the NCCN Compendium® (www.nccn.org).
¥Clinical validation demonstrated concordance with the following companion diagnostics: cobas® EGFR Mutation Test. Ventana ALK (DSF3) CDx Assay, Vysis ALK BreakApart FISH Probe Kit, therascreen® KRAS RGQ PCR Kit, Dako HER2 FISH PharmDx® Kit, cobas® BRAF V600 Mutation Test, THxlD® BRAF kit. For more information, please see the FoundationOne®CDx Technical Specifications available at: www.rochefoundationmedicine.com/f1cdxtech.
¶Clinical validation demonstrated concordance with the following diagnostics: cobas® EGFR Mutation Test v2, a tumor tissue polymerase chain reaction-based clinical trial assay (CTA), and an externally validated circulating cell-free DNA-based next-generation sequencing assay. For more information please see the FoundationOne Liquid®CDx Technical Specifications available here.
bTMB, blood Mutational Tumour Burden; FDA, US Food and Drug Administration; MSI, Microsatellite Instability; NCCN, Nat ional Comprehensive Cancer Network; NGS, next generation sequencing; TMB, Tumour Mutational Burden.
- FoundationOne CDx Technical Information (FDA Label). Available at: https://www.foundationmedicine.com/test/foundationone-cdx (Accessed May 2021).
- Data on file: FoundationOne Liquid CDx Technical Specifications, 2020.
- FoundationOne®Heme Technical Specifications, 2017. Available at: www.foundationmedicine.com/genomic-testing/foundation-one-heme (Accessed August 2020).
- Frampton GM et al. Nat Biotechnol 2013; 31: 1023–1031.
- Woodhouse et al. (2020) Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 15(9): e0237802. https://doi.org/10.1371/journal.pone.0237802.
- Clark TA et al. J Mol Diagn 2018; 20: 686–702.
- He J et al. Blood 2016; 127: 3004–3014.
- Suh JH et al. Oncologist 2016; 21: 684–691.
- Chalmers ZR et al. Genome Med 2017; 9: 34.
- Rozenblum AB et al. J Thorac Oncol 2017; 12: 258–268.
- Schrock AB et al. Clin Cancer Res 2016; 22: 3281–3285.
- Ross JS et al. Gynecol Oncol 2013; 130: 554–559.
- Hall MJ et al. J Clin Oncol 2016; 34: 1523–1523.
- FoundationOne®CDx Sample Report. F1CDx Lung High TMB_M-NZ-0257.pdf
- A search for "Foundation Medicine"[Affiliation] on the NCBI database 614 results at May 2021. Available at: https://www.ncbi.nlm.nih.gov/pubmed/?term=%22Foundation+Medicine%22%5BAffiliation%5D (Accessed August 2020).
- Foundation Medicine Publications. Available at: https://www.foundationmedicine.com/resources (Accessed August 2020). You have to change the third dropdown box to publications.
- FoundationOne®CDx FDA Approval, 2017. Available at:
https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019a.pdf (Accessed August 2020). - FoundationOne®CDx Clinical Validation, 2017. Available at: www.rochefoundationmedicine.com/f1cdxtech
- FoundationOne Liquid CDx FDA Approval, 2020. Available at: https://www.foundationmedicine.com/press-releases/445c1f9e-6cbb-488b-84ad-5f133612b721 (Accessed August 2020).